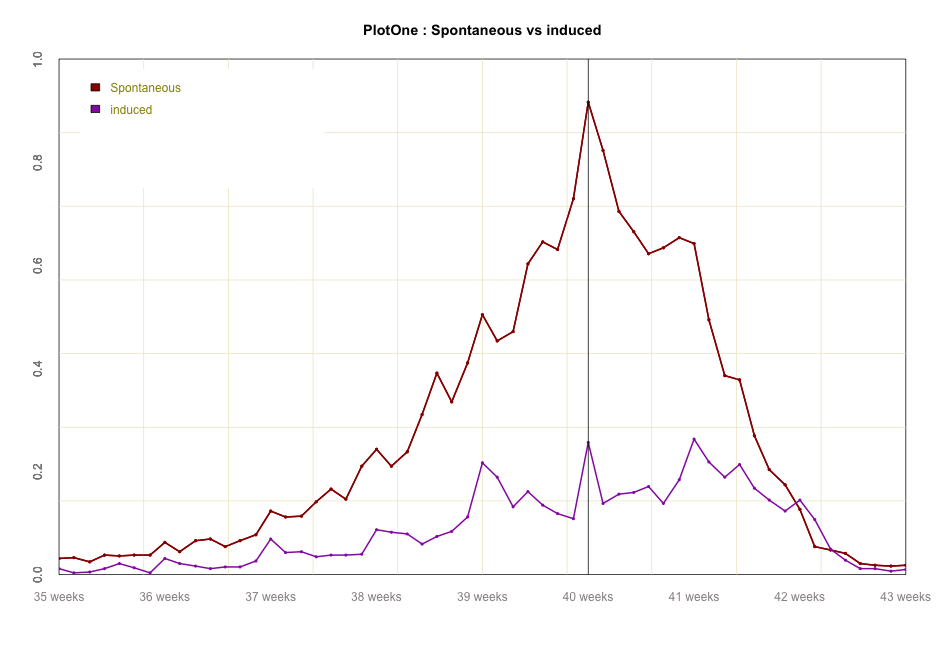
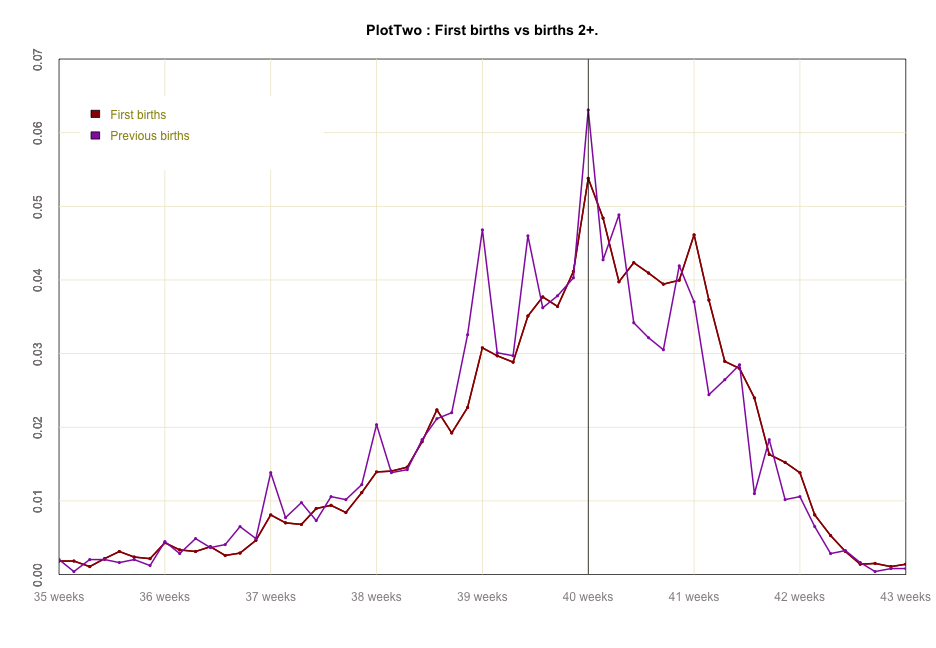
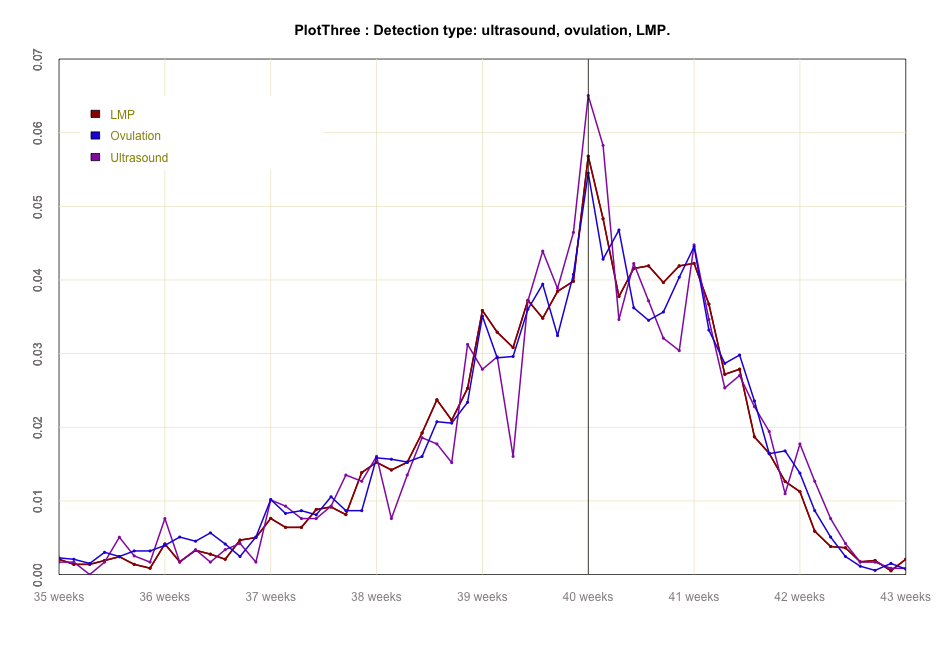
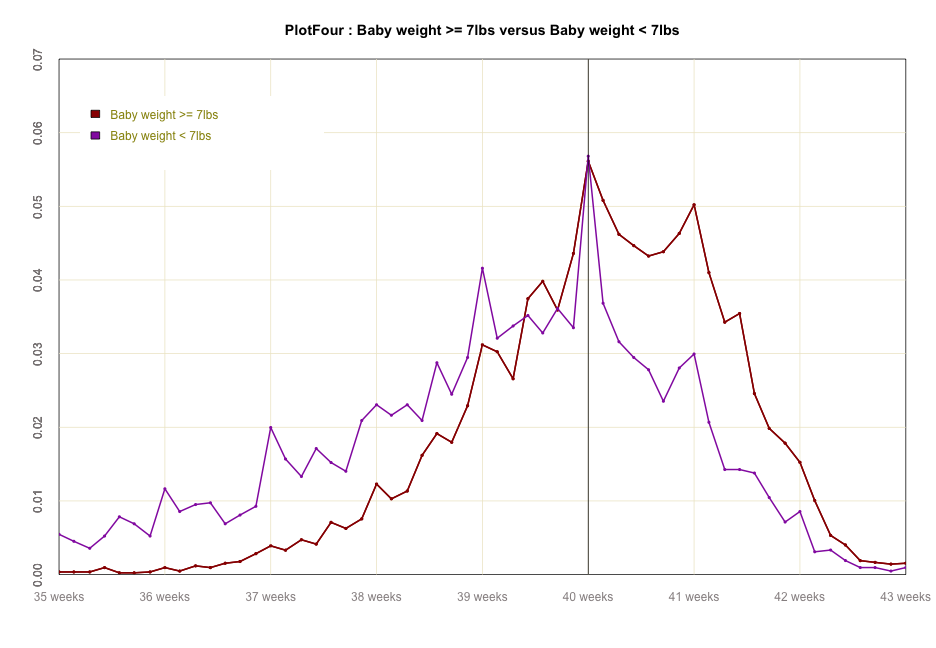
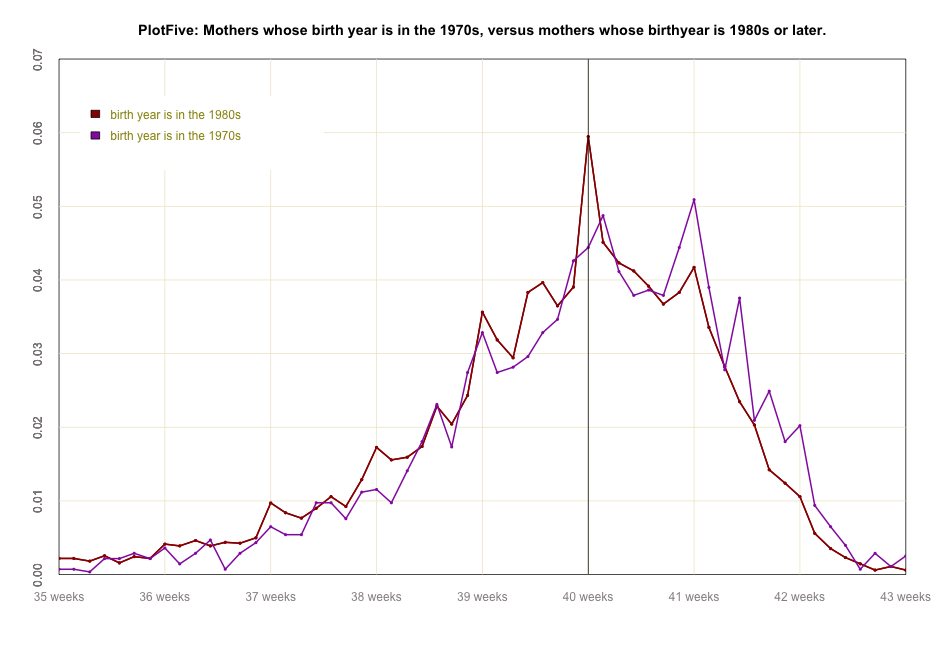
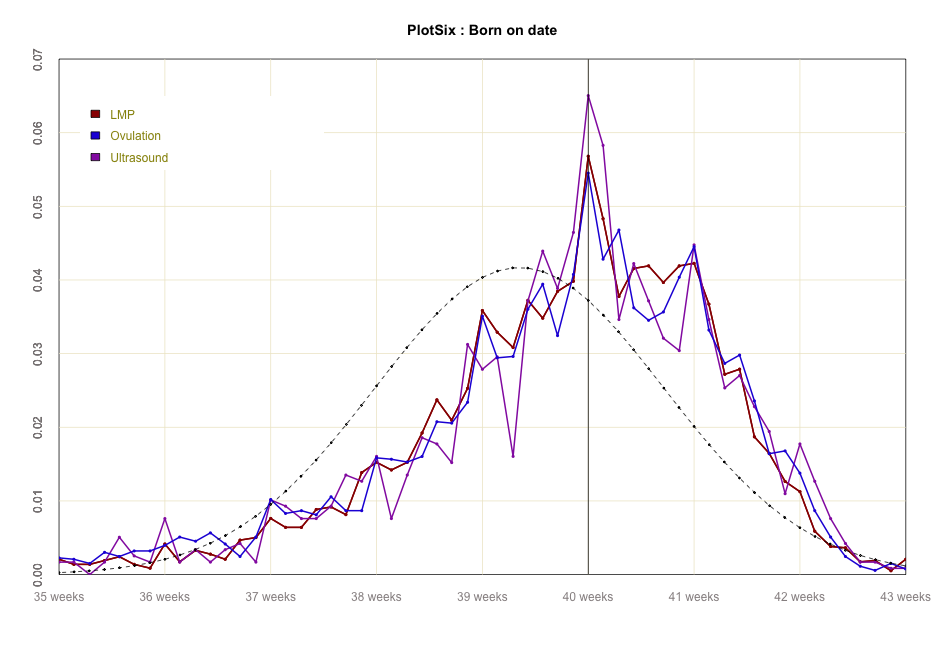
#global code
##global adjustments starts 01
rm(list=ls())
#seed for replication
set.seed(100)
oldwarnval <- getOption("warn") #preserve old value
options(warn = -1) #warnings are silent now
##global adjustments ends 01
#####generalized plot function
plotFunction <- function(x1,y1,x2,y2,x3 = -1,y3
,ztitle = "plot"
,zlegendText = c("var1","var2")
,customYlim = c(0,7/100)
,customLegend = c(35.2,37.5,0.065, 0.055)
,customGrid = c(8,7)
){
#X fisrt line plot
#Y second line plot
#Z thir line plot
x = x1;y = y1;
plot(x,y
,type= "o"
,main= ztitle
,col = rgb(6/10, 0, 0) #36,137,371 #color of the line chart
,lwd = 2 #line thickness
,pch = 19 # add points, solid circle,
,cex = 0.3 # size of the point on the chart
,tick = F
,xaxs = "i"
,yaxs = "i"
,xaxt = "n"
,ylim = customYlim
,xlab = ""
,ylab = ""
) #customGrid = c(8,7)
grid(nx = customGrid[1]#8
,ny = customGrid[2]#7
, lty=1, col = "cornsilk2")
lines(x, y
,type= "o"
,col = rgb(6/10, 0, 0) #36,137,371 #color of the line chart
,lwd = 2 #line thickness
,pch = 19 # add points, solid circle,
,cex = 0.3 # size of the point on the chart
)
x = x2; y = y2;
lines(x, y
,type= "o"
,col = rgb(149/255, 41/255, 176/255)
,lwd = 2
,pch = 19
,cex = 0.3
)
if(x3 != -1){
x = x3; y = y3;
zcol <- c((rgb(6/10, 0, 0)), rgb(36/255, 29/255, 219/255),
rgb(149/255, 41/255, 176/255)) #3 colors
zcol1 <- rgb(36/255, 29/255, 219/255)
lines(x, y, type = "o"
,col.lab="blue"
,col = zcol1
,lwd = 2
,pch = 19
,cex = 0.3
)
}else{
x = 0; y = 0;
zcol <- c((rgb(6/10, 0, 0)), rgb(149/255, 41/255, 176/255)) # 2 colors
}
axis(side = 1, at = 35:43
,labels = paste(c(35:43),"weeks")
,col.axis = rgb(151/255, 145/255, 145/255)
,tick = F
)
axis(side = 2
,col.axis = rgb(151/255, 145/255, 145/255)
,tick = F
)
legend( x = c(customLegend[1],customLegend[2] )
,y = c(customLegend[3],customLegend[4] ) #x=c(35.2,37.5),y=c(0.98, 0.75)
,legend = zlegendText
,fill = zcol
,text.col = rgb(151/255, 145/255, 10/255)
,border = 1
,box.lty= 0
,bty = "o"
,y.intersp=1.5
)
abline(v = 40)
#return(1)
}
#read file
df <- data.frame(read.csv("~/Downloads/Tony/Project 1/sf_gestationdata.csv",stringsAsFactors = F))